|
I am a second year PhD candidate at AMLab and AI4Science Lab at University of Amsterdam. My PhD project is about using deep learning tools for protein stabilization and peptide design. I work with Dr.Patrick Forré. I also collaborate with Janssen Vaccine Design. Prior to that, I obtained my Master's degree from University College London in Data Science and Machine Learning, and Bachelor's joint degree from UESTC (电子科技大学) and University of Glasgow in Communication Engineering. |

|
|
[Jan 2026] Our paper lead by Olga Zaghen on Riemmanian Variational Flow Matching was accepted by ICLR 2026! [Aug 2025] Our paper (with ByteDance Seed AI4Science) on Protein Mini-Binder Design, PXDesign was published! [Apr 2025] Our paper on Clifford Diffusion Models was accepted by ICLR 2025 FPI workshop. [Dec 2024] I will be joining ByteDance Seed AI4Science team as a research scientist intern! [Aug 2024] I am happy to give a presentation related to Clifford Neural Nets and Message Passing Simplicial Networks at AGACSE2024. [Jun. 2024] Our paper on Faster and Better Clifford GNNs was accepted by ICML 2024 GRaM workshop. [Feb. 2024] Our paper "Clifford Group Equivariant Simplicial Message Passing Networks" was accepted by ICLR 2024! |
|
I'm interested in AI4Science, specifically Protein Design and Generative Modeling. |
|
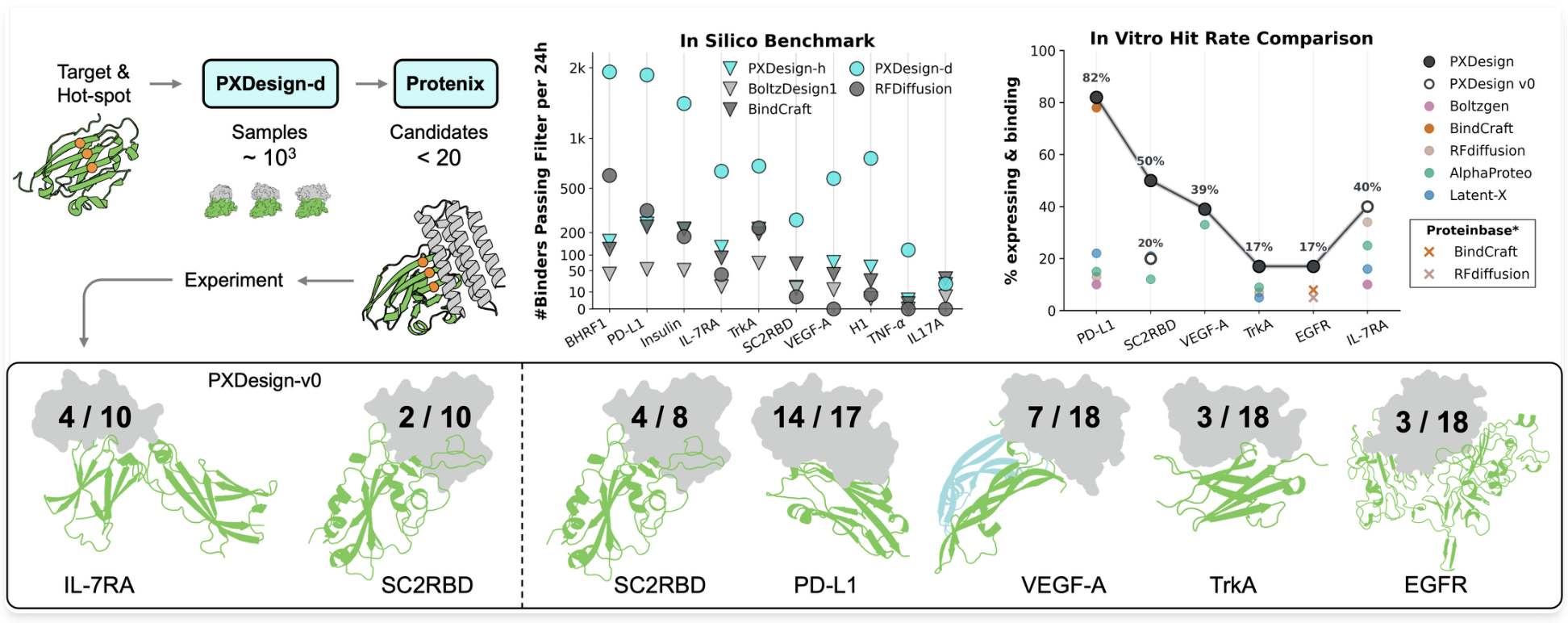
Milong Ren, Jinyuan Sun, Jiaqi Guan, Cong Liu, Chengyue Gong, et al. Technical Report, 2025 project page PXDesign is a fast and modular framework for de novo protein binder design. I contributed as a core team member, focusing on PXDesign-h development, evaluation pipeline and analysis. |
|
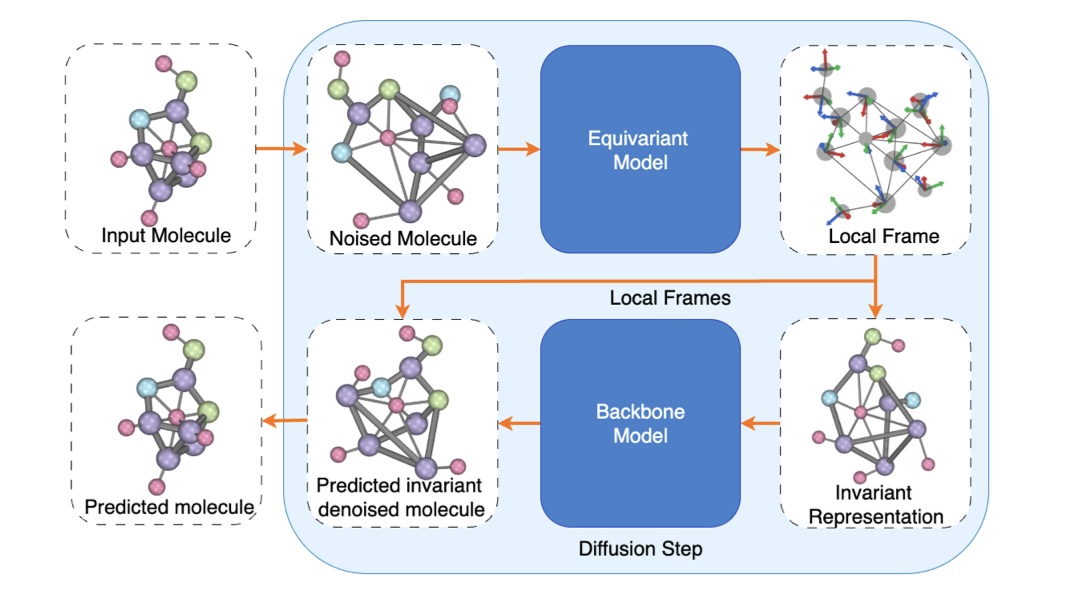
Mohan Guo*, Cong Liu*, Patrick Forré NeurIPS Workshop, 2025 paper We propose a frame-based diffusion paradigm for 3D molecular generation that achieves deterministic E(3)-equivariance while decoupling symmetry handling from the model backbone. The framework supports global and local frame constructions, improves scalability and sampling efficiency, and achieves state-of-the-art performance on QM9. |
|
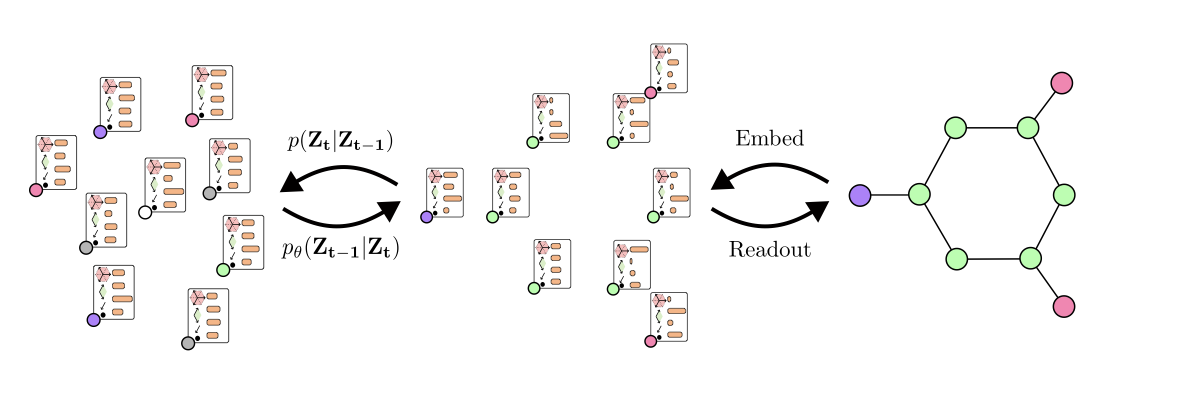
Cong Liu*, Sharvaree Vadgama*, David Ruhe, Erik Bekkers, Patrick Forré ICLR 2025 FPI Workshop paper We introduce Clifford Diffusion Models (CDMs), which leverage the expressive power of Clifford algebra for deterministic E(n)-equivariant diffusion. By extending diffusion beyond vector features to higher-grade multivector subspaces, the model captures joint geometric distributions across subspaces and enables expressive and physically grounded 3D molecular generation on QM9. |
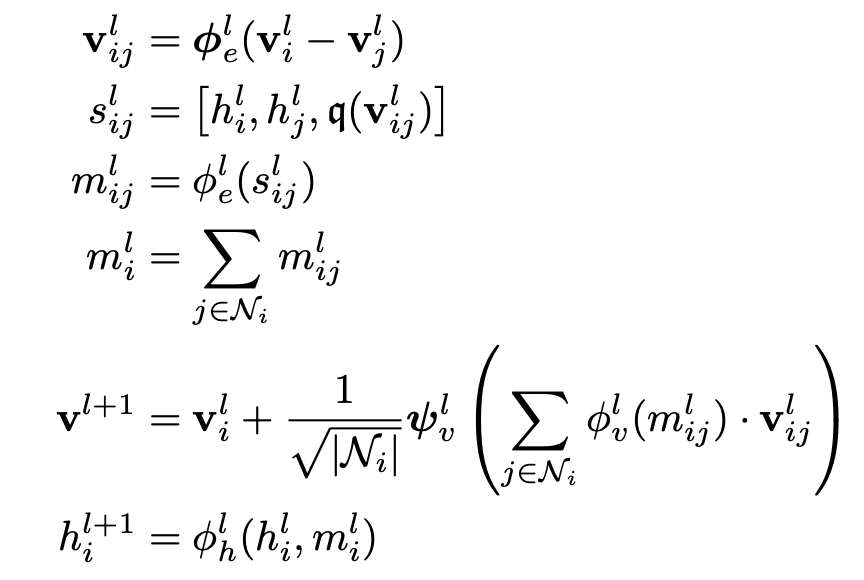
|
Cong Liu, David Ruhe, Patrick Forré, ICML GRaM workshop,2024 paper / code This paper focuses on faster and performance-wise better Clifford GNNs. |
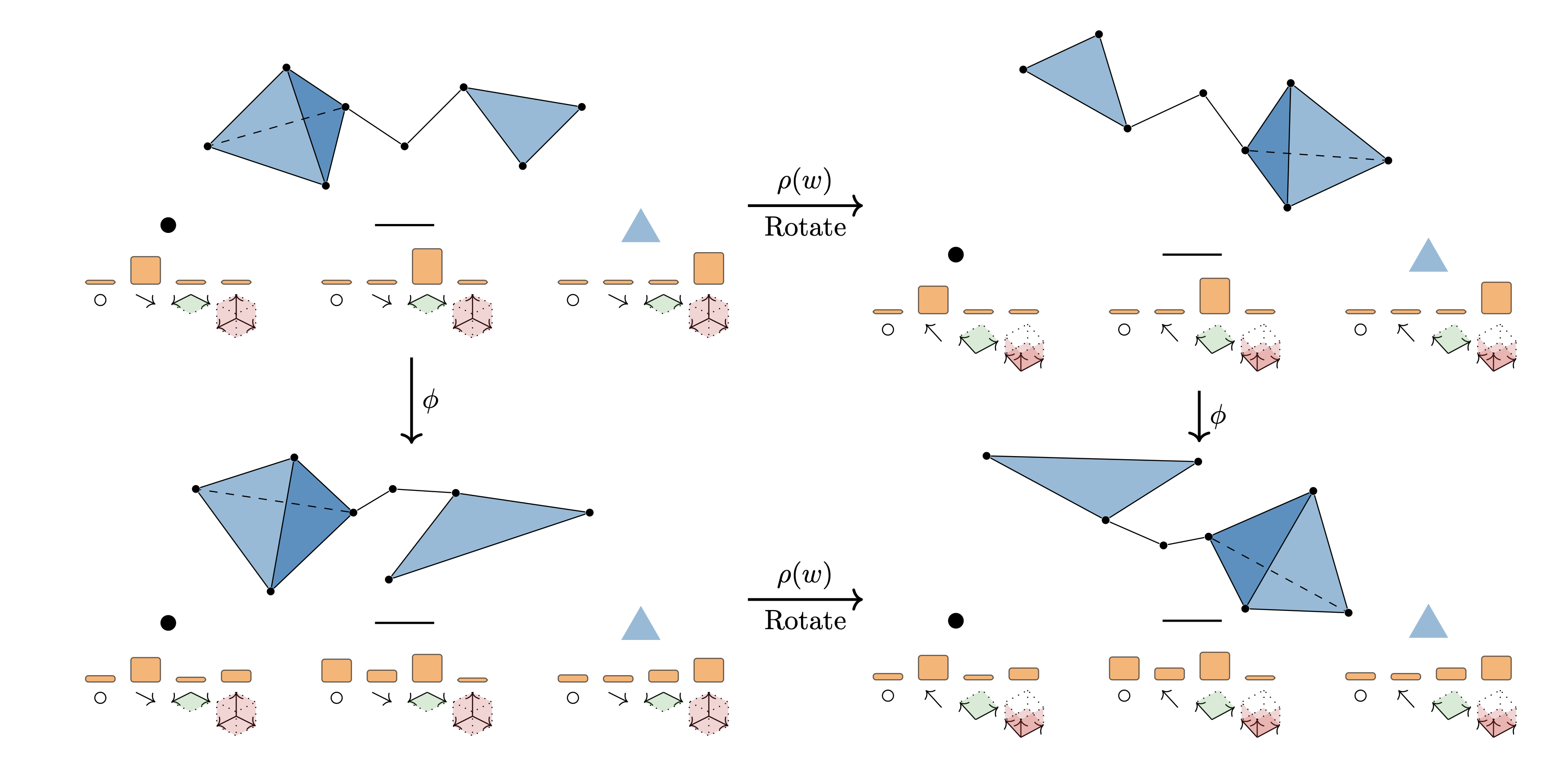
|
Cong Liu*, David Ruhe*, Floor Eijkelboom, Patrick Forré, ICLR, 2024 paper / code This paper proposes a general framework that considers both topological and geometric informatiom in general geometric graphs by leveraging Clifford group equivariant networks and simplicial message passing networks. |